Introduction
scEChIA (Single Cell Epigenome Chromatin Interaction Analysis) is a computational method that can predict chromatin interactions among distal sites with high accuracy. scEChIA can also make UCSC track on predicted interactions that may be useful for genomics studies based on chromatin interactions using the UCSC Genome Browser.
Installation
Download Package
or use the link http://reggen.iiitd.edu.in:1207/scEChIA/scEChIA_Package/scEChIA_0.1.0.tar.gz
Library required for scEChIA package:
Chromatin interaction prediction using prior rho information:
Function_name: Interaction_Prediction_1
How to run:
setwd(system.file("data", package="scEChIA")) #Keep your input data format like example data
genomic_region1 = read.table('K562_HiC_chr17.txt')
genomic_region1[is.na(genomic_region1)] = 0
genomic_region2 = read.table('IMR90_HiC_chr17.txt')
genomic_region2[is.na(genomic_region2)] = 0
gap = 25000 #Bin size 25kb (User can change it as per his choice)
patternf = 17 #Chromosome number (User can change it according to chromosome number)
chrNo = patternf
data = read.table('GM12878_SC_chr17.txt')
chrinfo = data[, 1:3] #Chromosome location (chr, start, end)
chrNo = patternf
startCell = 96 #start sample column
endCell = 575 #end sample column
chromSize = 83257441 #Size of chromosome 17 (Change it according to chromosome number. Follow heading Chromosome Size of hg19)
rhomatrix = rhomatAvg(genomic_region1, genomic_region2, gap, patternf, data, chrinfo)
predicted_interaction = Interaction_Prediction_1(chrinfo, data, rhomatrix, chrNo, startCell, endCell, chromSize)
Chromatin interaction prediction using constant rho:
Function_name: Interaction_Prediction_2
How to run:
gap = 25000 #Bin size 25kb (User can change it as per his choice)
patternf = 17 #Chromosome number (User can change it according to chromosome number)
chrNo = patternf
data = read.table('GM12878_SC_chr17.txt')
chrinfo = data[, 1:3] #Chromosome location (chr, start, end)
chrNo = patternf
startCell = 96 #start sample
endCell = 575 #end sample
chromSize = 83257441 #Size of chromosome 17 (Change it according to chromosome number. Follow heading Chromosome Size of hg19)
rhoConstant = 0.01 #user can change the value of rhoConstant
predicted_interaction = Interaction_Prediction_2(chrinfo, data, rhoConstant, chrNo, startCell, endCell, chromSize)Visualizing chromatin interactions using UCSC track
Function_name: ucscTrack
How to Run :
predInteraction = predicted_interaction
addscore = 500 #User can increase or decrease addscore value as per his choice
ucscTrack_file = ucscTrack(predInteraction, addscore)
Note :
After generating the UCSC track file, the bottom line has to be pasted on the header to be visualized on the UCSC Genome Browser. track type=interact name="User and celltype" description="User and celltype" interactDirectional=true maxHeightPixels=200:100:50 visibility=full
#chrom chromStart chromEnd name score value exp color sourceChrom sourceStart sourceEnd sourceName sourceStrand targetChrom targetStart targetEnd targetName targetStrand
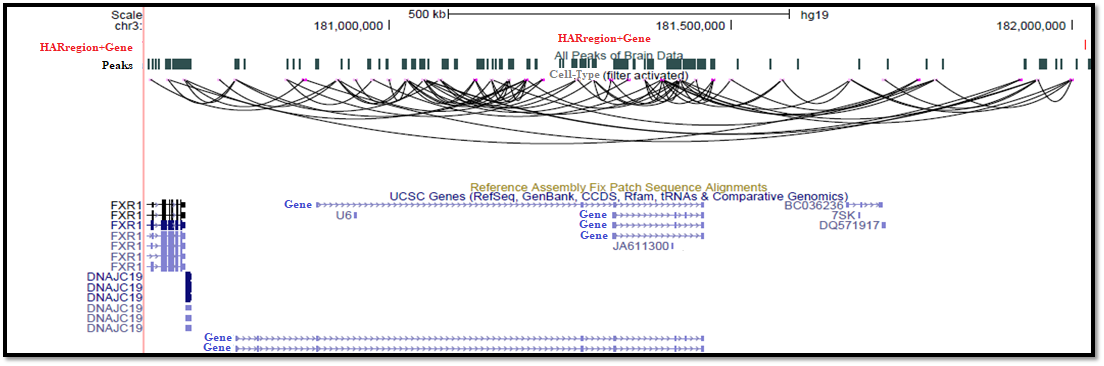
Visualization figure of microglia using UCSC Genome Browser (https://genome.ucsc.edu/) :
How to validate predicted chromatin interactions? :
1. Predict the chromatin interaction using scATAC-seq data from scEChIA. 2. Find the related available chromatin interaction from HiC/chromatin-based data and make it paired for every interaction. 3. Use freely available resource - paired genomic loci tool ( pgltools - https://github.com/billgreenwald/pgltools) to intersect predicted chromatin interactions with existing chromatin interactions. 4. Check the fraction/number of overlap.
Chromosome Size of hg19
chr1: 248956422, chr2: 242193529, chr3: 198295559, chr4: 190214555, chr5: 181538259, chr6: 170805979, chr7: 159345973, chr8: 145138636, chr9: 138394717, chr10: 133797422, chr11: 135086622, chr12: 133275309, chr13: 114364328, chr14: 107043718, chr15: 101991189, chr16: 90338345, chr17: 83257441, chr18: 80373285, chr19: 58617616, chr20: 64444167, chr21: 46709983, chr22: 50818468, chrX: 156040895, chrY: 57227415