ePerturbDB API Documentation
Overview
The ePerturbDB API provides programmatic access to enhancer perturbation data stored in our SQLite database. All responses are returned in JSON format.
- Database: SQLite
- Server: Apache (Ubuntu)
- Base URL:
http://reggen.iiitd.edu.in:1207/ePerturbDB-html/api/
API Reference
The ePerturbDB API provides access to enhancer perturbation data through RESTful endpoints. Below are the details of available endpoints and their parameters.
Endpoints
1. Get Enhancers
GET /enhancers.php?limit=3
Retrieve enhancer records. Optional parameter:
limit– Number of results (default: 50)
2. Get Enhancer by ID
GET /enhx-id.php?id=enhrx-106476
Retrieve enhancer details by Enhancer-id.
3. Get Enhancers by Gene Name
GET /gene-id.php?gene=APP
Retrieve enhancers associated with a specific gene (matches Gene (literature) or nearest_gene_name).
4. Pathway Enrichment
GET /pathway_enrichment.php?gene=APP
Retrieve pathway enrichment data for a given gene.
Usage Examples
cURL Examples
1. Get all enhancers (default limit: 50):
curl "http://reggen.iiitd.edu.in:1207/ePerturbDB-html/api/enhancers.php"
2. Get enhancers by gene name:
curl "http://reggen.iiitd.edu.in:1207/ePerturbDB-html/api/gene-id.php?gene=APP"
3. Get enhancer by ID:
curl "http://reggen.iiitd.edu.in:1207/ePerturbDB-html/api/enhx-id.php?id=enhrx-106476"
Python Example
import requests
url = "http://reggen.iiitd.edu.in:1207/ePerturbDB-html/api/gene-id.php"
params = {"gene": "APP"}
response = requests.get(url, params=params)
data = response.json()
print(data)
Frequently Asked Questions
No, the current version is open access.
The backend uses SQLite for lightweight and fast access.
Currently, there are no strict rate limits implemented. However, we recommend limiting requests to 100 per minute to ensure optimal performance for all users.
The database is updated quarterly with new enhancer perturbation data from published literature and experiments.
All API responses are returned in JSON format. Each enhancer record includes fields such as enhancer ID, genomic coordinates, associated genes, and perturbation data.
Please report any issues or bugs to the REGGEN Lab team via email. Include details about the endpoint, parameters used, and the error encountered.
Yes, you can download the complete dataset in Excel format from our Downloads page. For programmatic bulk access, you can use the enhancers.php endpoint with appropriate limit parameters.
Please cite our database as: "REGGEN Lab (2025). ePerturbDB: A database of enhancer perturbations. Available at: http://reggen.iiitd.edu.in:1207/ePerturbDB-html/"
To visualise our enhancer BED detail files in UCSC:
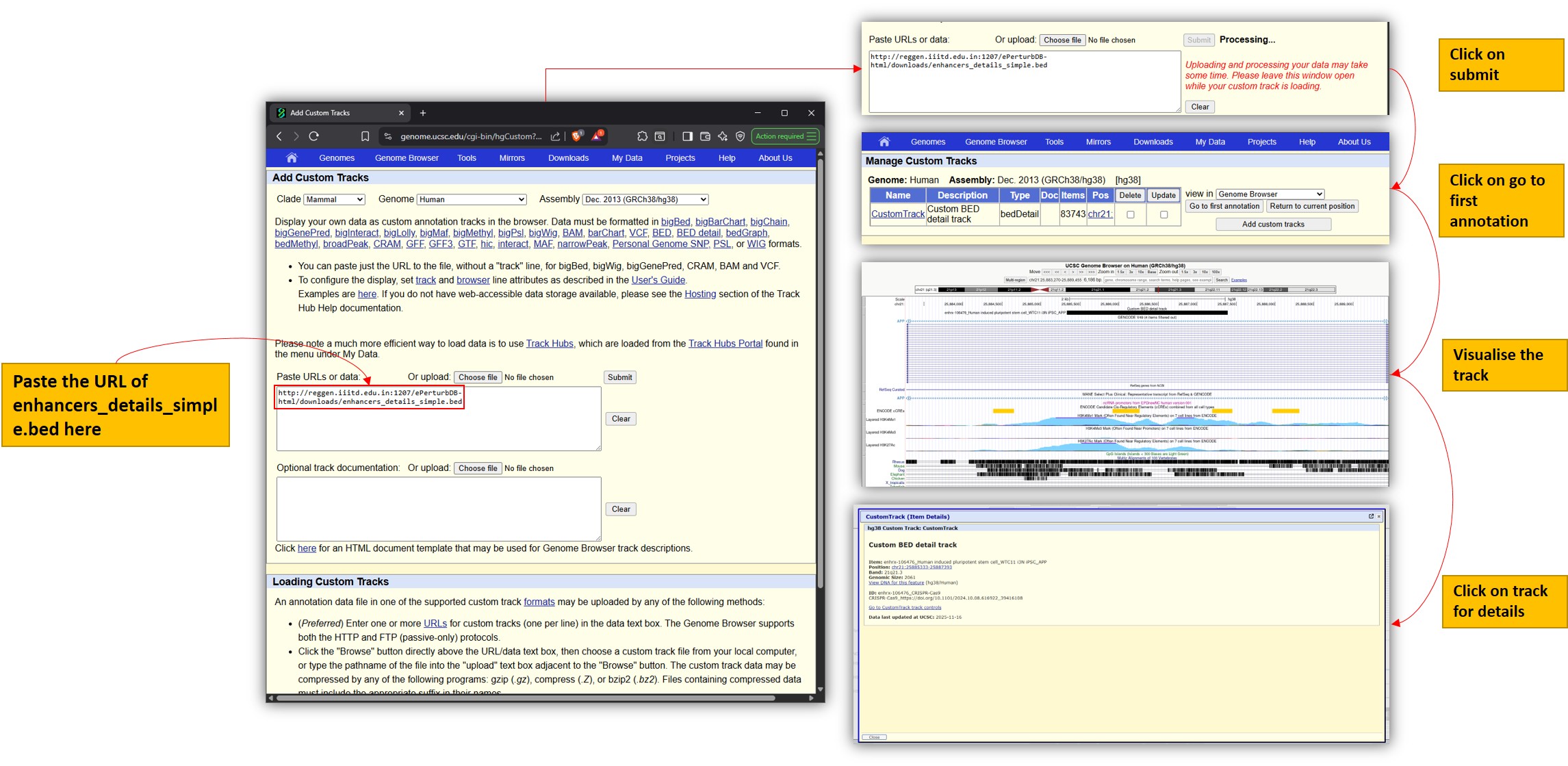
- Open the UCSC Genome Browser and go to "Add Custom Tracks".
- Paste the URL of the BED detail file from our Downloads page.
- Submit, then click "go to first annotation" to load the track.
- Click on the track to view enhancer details.
